Blood
Pressure Database Example
Tasks covered:
Working
with .RData files [save and load]
Creating
and working with functions
Working
with date data types
Working
with date sequences
Plotting
multiples data series on a single chart
Annotating
a chart axis with dates and and displaying the dates perpendicular to
the axis
This
example R script demonstrates how to use an R data frame as a database.
It includes code to create an initial blood pressure database. and two
functions, one to add new blood pressure entries to the database, and
another to chart the data in the database. Code to create two example
52-week blood pressure databases is also included to help demonstrate
the use of the two included functions.
This
example demonstrates several interesting skills with R. The code shows
how to use dates in a data frame, how to show dates along a chart axis,
how to use date data to position a chart legend, how to plot multiple
lines [data and reference] on the same chart, and the use of color, line
type, and line width in a chart.
Create
an initial blood pressure database
The
first block of code can be used to create an initial entry in your
database.
##########################################################################
#
use this code to create your own bp database - skip this if you are
simply going to run the example
#
add new data entries with add_bp_entry( )
#
chart the database with view_bp_chart( )
#
create the blood pressure database as a data frame [use this code to
create a new database]
bp_db
<- as.data.frame(cbind(0, 0, 0)) # create the initial database
entry
bp_db[,1]
<- Sys.Date() # insert the current date into the first column
names(bp_db)
<- c('Date','Systolic', 'Diastolic') # assign column titles
save(bp_db,
file = 'bp_db.RData') # save the database
#
end of the creation your initial database
##########################################################################
The
code
creates an initial entry in a data frame containing zeros for all of the
values. The current date is inserted into the first column. Column
titles are assigned to the data frame, and the data frame is saved as an
.RData file. This data frame can be used as a blood pressure database
with the functions add_bp_entry( ) and view_bp_chart( ).
Example
blood pressure databases
The
next block of code creates two example blood pressure databases. These
are helpful in testing the functionality of the two project functions
that will be discussed below.
##########################################################################
# use this code to create the example bp databases
# create the example databases [these are used in the example below]
# add new data entries with add_bp_entry( )
# chart the database with view_bp_chart( )
# create example data
dateSeq <- seq(as.Date("2017-06-03"), by = "week", length.out = 52) #
a sequence of 52 weekly dates
sysSeq <- seq(120, 138) # a set of systolic bp values
diaSeq <- seq(68, 85) # a set of distolic bp values
# a function to create our example bp sequences
sampleSeq <- function(s, x) {
r <- c() # an empty list
for (i in 1:x) { # repeat x times
r <-rbind(r, sample(s, 1)) # add a random value
from s to list r
}
r # return list r with x elements
}
# create bob's bp database [the dates will be inserted incorrectly]
# generate example bp values
s <- sampleSeq(sysSeq, 52) # a sequence of 52 random systolic bp
values
d <- sampleSeq(diaSeq, 52) # a sequence of 52 random diastolic bp
values
bob_bp_db <- as.data.frame(cbind(dateSeq,s,d)) # combine the data
into a dataframe
# cbind( ) coerces the dates to integer values
bob_bp_db[,1] <- dateSeq # fix the dates in the first column
names(bob_bp_db) <- c('Date','Systolic','Diastolic') # assign column
titles
save(bob_bp_db, file = 'bob_bp_db.RData') # save the database
# create carol's bp database [the dates will be inserted incorrectly]
# generate example bp values
s <- sampleSeq(sysSeq, 52) # a sequence of 52 random systolic bp
values
d <- sampleSeq(diaSeq, 52) # a sequence of 52 random diastolic bp
values
carol_bp_db <- as.data.frame(cbind(dateSeq,s,d)) # combine the data
into a dataframe
# cbind( ) coerces the dates to integer values
carol_bp_db[,1] <- dateSeq # fix the dates in the first column
names(carol_bp_db) <- c('Date','Systolic','Diastolic') # assign
column titles
save(carol_bp_db, file = 'carol_bp_db.RData') # save the database
# end of the creation the example databases
###########################################################################
The
first three lines of the code generate three sequences: a weekly date
sequence spanning 52 weeks, a sequence of systolic blood pressure
values from 120 to 138, and a sequence of diastolic blood pressure
values from 68 to 85.
# create example
data
dateSeq <- seq(as.Date("2017-06-03"), by = "week", length.out = 52) #
a sequence of 52 weekly dates
sysSeq <- seq(120, 138) # a set of systolic bp values
diaSeq <- seq(68, 85) # a set of distolic bp values
The
next section of code defines a function that we will use to generate the
series
of systolic and diastolic values for each weekly reading in
our example databases.
# a function to
create our example bp sequences
sampleSeq <- function(s, x) {
r <- c() # an empty list
for (i in 1:x) { # repeat x times
r <-rbind(r, sample(s, 1)) # add a random value
from s to list r
}
r # return list r with x elements
}
The
function
sampleSeq has two input arguments: a
sequence of values and the desired number of values in
the output list. The first line of code creates an empty list, names r,
where the generated values will be stored. Then the function loops for x
iterations. In each loop, the input sequence is randomly sampled for and
value and that value is inserted in the output list. After the loop is
complete, the output list r of x elements is returned.
The
next two code blocks create the two example blood pressure databases.
Both blocks are identical with the exception that one creates a database
called bob_bp_db
and the other creates a database called carol_bp_db, therefore only the first block
will be described here.
# create bob's bp
database [the dates will be inserted incorrectly]
# generate example bp values
s <- sampleSeq(sysSeq, 52) # a sequence of 52 random systolic bp
values
d <- sampleSeq(diaSeq, 52) # a sequence of 52 random diastolic bp
values
bob_bp_db <- as.data.frame(cbind(dateSeq,s,d)) # combine the data
into a dataframe
# cbind( ) coerces the dates to integer values
bob_bp_db[,1] <- dateSeq # fix the dates in the first column
names(bob_bp_db) <- c('Date','Systolic','Diastolic') # assign column
titles
save(bob_bp_db, file = 'bob_bp_db.RData') # save the database
The
function sampleSeq( )
is used to generate two 52 element sequences, one for the systolic blood
pressure values and another for the diastolic values. These two
sequences are combined with the weekly dates into the data frame bob_bp_db
using cbind( ).
Unfortunately, cbind( )
coerces the date values into integer values before as.data.frame(
) creates our data
frame, so the date sequence is reassigned to the first column of bob_bp_db.
Now, the first column of the data frame is populated correctly with our
sequence of weekly dates. The column titles of Date, Systolic, and
Diastolic are added to the data frame. finally the data frame is saved
as an RData file. This process is repeated for carol_bp_db.
Manage
the blood pressure databases [add and view the data]
The
next block of code creates the two functions for the blood pressure
database. The function add_bp_entry(
) adds a new entry [current date, systolic BP,
diastolic BP] to the database and saves it. The function view_bp_chart(
) charts the data in a selected database.
##########################################################################
# the bp database functions
add_bp_entry <- function(DB, systolic, diastolic) { # add an entry
to the db
if(DB == 'bob') {
load('bob_bp_db.RData') # open the database
new_row <- as.data.frame(cbind(0, systolic,
diastolic)) # create a new row
new_row[,1] <- Sys.Date() # insert the current
date in the first column
names(new_row) <- c('Date','Systolic',
'Diastolic') # add column titles
result <- rbind(bob_bp_db, new_row) # add the
new row to the database
bob_bp_db <- result
save(bob_bp_db, file = 'bob_bp_db.RData') # save
the database
} else {
load('carol_bp_db.RData') # open the database
new_row <- as.data.frame(cbind(0, systolic,
diastolic)) # createa a new row
new_row[,1] <- Sys.Date() # insert the current
date in the first column
names(new_row) <- c('Date','Systolic',
'Diastolic') # add column titles
result <- rbind(carol_bp_db, new_row) # add the
new row to the database
carol_bp_db <- result
save(carol_bp_db, file = 'carol_bp_db.RData') #
save the database
}
}
view_bp_chart <- function(DB) { # chart the bp data
if(DB == "bob") {
load('bob_bp_db.RData') # open the database
startDate <- bob_bp_db[1,1] # get the first date
in the database to position the legend correctly
# plot a line chart of the systolic bp [note that
xaxt='n' supresses the x-axis values]
plot(bob_bp_db$Systolic ~ bob_bp_db$Date,
col='red', type = 'l', ylim = c(50,145), xaxt='n',
main =
'Bob\'s Blood Pressure', xlab = 'Date', ylab = 'BP')
# add dates as the x-axis values perpendicular [las
= 2] to the axis
axis(1, bob_bp_db$Date, labels =
format(bob_bp_db$Date, "%b-%d"), las = 2, cex.axis = .7)
# add the diastolic bp line
lines(bob_bp_db$Diastolic ~ bob_bp_db$Date, col =
'green')
} else {
load('carol_bp_db.RData') # open the database
startDate <- carol_bp_db[1,1] # get the first
date in the database to position the legend correctly
# plot a line chart of the systolic bp [note that
xaxt='n' supresses the x-axis values]
plot(carol_bp_db$Systolic ~ carol_bp_db$Date,
col='red', type = 'l', ylim = c(50,145), xaxt='n',
main =
'Carol\'s Blood Pressure', xlab = 'Date', ylab = 'BP')
# add dates as the x-axis values perpendicular [las
= 2] to the axis
axis(1, carol_bp_db$Date, labels =
format(carol_bp_db$Date, "%b-%d"), las = 2, cex.axis = .7)
# add the diastolic bp line
lines(carol_bp_db$Diastolic ~ carol_bp_db$Date, col
= 'green')
}
grid() # show a grid
abline(h = 120, lty = 3, col = 'red') # normal systolic bp line
abline(h = 140, lty = 3, lwd = 2, col = 'red') # borderline
systolic bp line
abline(h = 80, lty = 3, col = 'green') # normal diastolic bp
line
abline(h = 90, lty = 3, lwd = 2, col = 'green') #
borderline diastolic bp line
# add a legend
legend(as.Date(startDate),115, c('Systolic','Diastolic'),lty =
c(1,1), col = c('red','green'))
}
# end of the database functions
##########################################################################
The function add_bp_entry(
) will add a daily
entry to the selected blood pressure database.
add_bp_entry <-
function(DB, systolic, diastolic) { # add an entry to the db
if(DB == 'bob') {
load('bob_bp_db.RData') # open the database
new_row <- as.data.frame(cbind(0, systolic,
diastolic)) # create a new row
new_row[,1] <- Sys.Date() # insert the current
date in the first column
names(new_row) <- c('Date','Systolic',
'Diastolic') # add column titles
result <- rbind(bob_bp_db, new_row) # add the new
row to the database
bob_bp_db <- result
save(bob_bp_db, file = 'bob_bp_db.RData') # save the
database
} else {
load('carol_bp_db.RData') # open the database
new_row <- as.data.frame(cbind(0, systolic,
diastolic)) # createa a new row
new_row[,1] <- Sys.Date() # insert the current
date in the first column
names(new_row) <- c('Date','Systolic',
'Diastolic') # add column titles
result <- rbind(carol_bp_db, new_row) # add the
new row to the database
carol_bp_db <- result
save(carol_bp_db, file = 'carol_bp_db.RData') # save
the database
}
}
This function has three input
arguments: the database to add the new entry to, the systolic blood
pressure value, and the diastolic blood pressure value. The function
is separated into two duplicate sections. The first section adds an
entry into bob_bp_db ,
and the second section adds an entry into carol_bp_db
. Since the second section is
essentially a mirror of the first section, so this description will
only cover the functionality of the first section.
The
first code line opens the database file for editing in this function.
Once add_bp_entry( ) ends, the blood pressure database is not active.
The next line of code, a new data frame is created containing 0, the
input systolic value, and the input diastolic value using as.data.frame( )
and cbind( ) .
The first value of this data frame is replaced with the current date
using Sys.Date( )
and the column titles [Date, Systolic, and Diastolic] are added with
the function names( ) .
It is necessary to have matching column titles so that new new data
frame can be added to the existing database. R will raise an error if
the column titles do not match. Finally, the new row is added to the
database with rbind( ) and the updated database is saved
to an RData file. These steps are repeated in the second section for
carol_bp_db. The command add_bp_entry('bob',
121, 72) will add a
new entry to the database bob_bp_db containing today's date,
a systolic blood pressure value of 121, and a
diastolic blood pressure value of 72.
It
is important to understand that the database file is open and only
available inside this function and not in your main R session. Even if
the database is active in your current R session, this instance of the
database will be opened, updated, and saved without altering the open
version in your main R session.
The function view_bp_chart(
) creates a line
chart of the systolic and diastolic blood pressure values in the
selected blood pressure database. The function consists of three
sections. The first section charts bob_bp_db. The second section charts carol_bp_db.
The third section adds common details to either chart. The details
of how the charts are defined and presented are worth reviewing and
understanding.
view_bp_chart
<- function(DB) { # chart the bp data
if(DB == 'bob') {
load('bob_bp_db.RData') # open the database
startDate <- bob_bp_db[1,1] # get the first date
in the database to position the legend correctly
# plot a line chart of the systolic bp [note that
xaxt='n' supresses the x-axis values]
plot(bob_bp_db$Systolic ~ bob_bp_db$Date,
col='red', type = 'l', ylim = c(50,145), xaxt='n',
main = 'Bob\'s Blood Pressure', xlab = 'Date', ylab
= 'BP')
# add dates as the x-axis values perpendicular [las
= 2] to the axis
axis(1, bob_bp_db$Date, labels =
format(bob_bp_db$Date, "%b-%d"), las = 2, cex.axis = .7)
# add the diastolic bp line
lines(bob_bp_db$Diastolic ~ bob_bp_db$Date, col =
'green')
} else {
load('carol_bp_db.RData') # open the database
startDate <- carol_bp_db[1,1] # get the first
date in the database to position the legend correctly
# plot a line chart of the systolic bp [note that
xaxt='n' supresses the x-axis values]
plot(carol_bp_db$Systolic ~ carol_bp_db$Date,
col='red', type = 'l', ylim = c(50,145), xaxt='n',
main = 'Carol\'s Blood Pressure', xlab = 'Date',
ylab = 'BP')
# add dates as the x-axis values perpendicular [las
= 2] to the axis
axis(1, carol_bp_db$Date, labels =
format(carol_bp_db$Date, "%b-%d"), las = 2, cex.axis = .7)
# add the diastolic bp line
lines(carol_bp_db$Diastolic ~ carol_bp_db$Date, col
= 'green')
}
grid() # show a grid
abline(h = 120, lty = 3, col = 'red') # normal systolic bp line
abline(h = 140, lty = 3, lwd = 2, col = 'red') # borderline
systolic bp line
abline(h = 80, lty = 3, col = 'green') # normal diastolic bp
line
abline(h = 90, lty = 3, lwd = 2, col = 'green') #
borderline diastolic bp line
# add a legend
legend(as.Date(startDate),115, c('Systolic','Diastolic'),lty =
c(1,1), col = c('red','green'))
}
The
function view_bp_chart( ) has one input argument that
identifies which blood pressure database (bob or carol) to chart. The
designated RData file is opened. The first date entry is saved so the
chart legend can be properly positioned later in the function. A chart
is plotted for the systolic blood pressure values with the command
plot(bob_bp_db$Systolic ~ bob_bp_db$Date, col='red', type = 'l', ylim
= c(50,145), xaxt='n',
main = 'Bob\'s Blood Pressure', xlab = 'Date', ylab = 'BP')
Let's
examine each argument of this plot( ) function call. The first argument
[bob_bp_db$Systolic ~ bob_bp_db$Date] designates bob_bp_db$Date as the x-axis and bob_bp_db$Systolic as the y-axis of our chart. This
argument uses the R model formula notation. The format for this
notation is response ~ predictor. For the plot(
) function, the
predictor defines the x-axis variable and the response defines the
y-axis variable. The second argument [col='red'] sets the color of the plot to
red. The third argument [type = 'l'] sets the plot type to line
[the default type is dot]. The fourth argument [ylim
= c(50,145)] sets the
limit values for the y-axis. R normally sets the axis limits
based on the maximum and minimum values of the data being
plotted. If you wish specific maximum and minimum
limits,
as we do here, you must designate them using the ylim [or xlim]
arguments. These limits will be maximum and minimum
values
in the ranges you want your chart to show. In this example we
are designating specific minimum and maximum values
for the y-axis so we can set the location for our
chart legend later. The
fifth argument [xaxt='n']
suppresses the default x-axis index values. We will add custom
labels in another command and we must suppress the default
axis values to prevent conflicting duplicate values. The sixth
argument [main = 'Bob\'s Blood
Pressure'] adds the
chart title. Notice how the apostrophe is designated using
the symbol set \'. The final two arguments [xlab
= 'Date', ylab = 'BP'>]
assign the labels for the x-axis and y-axis.
The
all of the remaining commands use functions that add content to
the current existing chart rather than creating a new chart. The
axis( )
command adds a customized x-axis to our chart.
axis(1, bob_bp_db$Date, labels =
format(bob_bp_db$Date, "%b-%d"), las = 2, cex.axis = .7)
The
first argument in the axis( ) function identifies which
side of the chart this function will customize. The possible
argument values are:
1 = below, 2 = left, 3 = above, and 4 = right. The second
argument [bob_bp_db$Date]
identifies the data that will define the axis index values. The
third argument [labels
= format(bob_bp_db$Date, "%b-%d")] defines the
format for the index value. This example uses a month-date
format. The fourth argument [las
= 2] defines the index label orientation in
relation to the axis. The argument values identify whether the labels are parallel (=0) or
perpendicular(=2) to axis. The final argument [cex.axis
= .7] is the magnification
of axis annotation relative to cex, where cex is the number indicating the amount
by which plotting text and symbols should be scaled relative to
the default. 1=default, 1.5 is 50% larger, 0.5 is 50% smaller,
etc. In this example, the axis index labels are 70% of normal
size.
The
next command adds another line plot to our chart. This
line will chart Bob's diastolic blood pressure.
lines(bob_bp_db$Diastolic ~ bob_bp_db$Date, col =
'green')
The
lines( )uses
two arguments in this example. The first argument [bob_bp_db$Diastolic ~
bob_bp_db$Date] uses the R model formula
notation, bob_bp_db$Diastolic
is identified as the response and bob_bp_db$Date is
identified as the predictor. Notice that this
function uses the same predictor variable as our plot(
) function above. This guarantees that this
added line plot will coincide with the line plot in the main
chart. The second argument [col
= 'green'] sets the color of the new line to
green. This completes the section of commands for Bob's
individual chart. The next six commands will add generic content
to either chart. This concludes the portion of the function
creating the main blood pressure chart. This code is repeated if
the other database is selected for charting.
The
final block of code adds common features to the existing blood
pressure chart. The function grid()
adds a light grid to the background of the chart area. The
function abline(
) is used four times. The first use inserts a
horizontal dotted red line at the maximum normal systolic blood
pressure value of 120. The
second use inserts a heavy horizontal dotted red line
at the borderline high systolic blood pressure value
of 140. The
third use inserts a horizontal dotted green line at the
maximum normal diastolic blood pressure value of 80. The fourth
use inserts a heavy horizontal dotted green line
at the borderline high diastolic blood pressure
value of 80. These reference lines enhance a
viewer's understand what the chart values can
mean in the context of normal and high blood
pressure values.
The
final command adds a legend to the chart so the viewer knows
what the charted lines represent.
legend(as.Date(startDate),115,
c('Systolic','Diastolic'),lty = c(1,1), col = c('red','green'))
The
first two arguments of legend(
) [as.Date(startDate),115]
identify the x-y location of the upper left corner of the legend
box [as.Date(startDate),115].
These values are expressed in terms of the x and y axis values.
In this example, the x coordinate is a date saved when the
database was loaded at the beginning of the function view_bp_chart( )[the first
date in the selected database]. The y coordinate is a blood
pressure value. The value 115 was chosen so the legend is well
placed vertically on the chart. The third argument [c('Systolic','Diastolic')]
designates the text labels for the legend items. These labels
are stored in a list created with the function c(
). The fourth argument [lty
= c(1,1)] designates that the legend items
should be shown as lines. The final argument [col
= c('red','green')] designates the color for
each legend item.
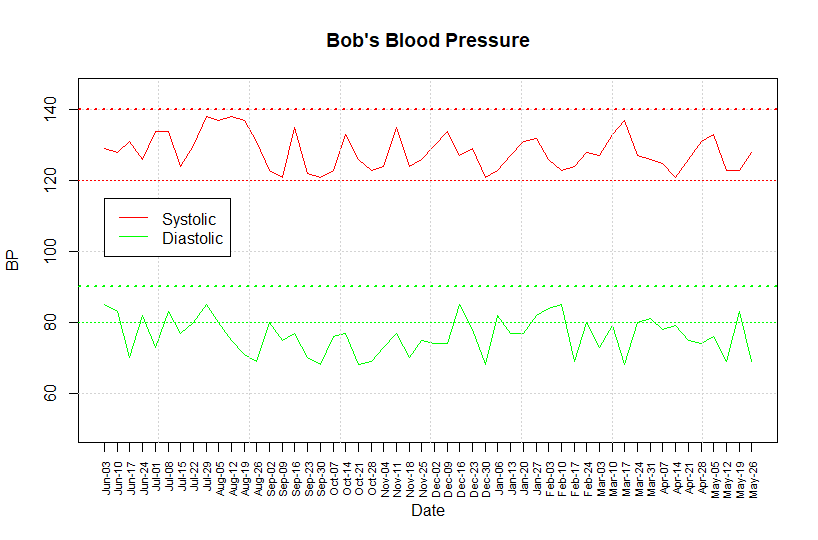
Here
is what the chart looks like for the example database bob_bp_db.RData.
This
little project is a good demonstration of several very useful R
skills and techniques.